Abstract
Although follicular lymphoma (FL) usually has an indolent clinical course, up to 20% of patients experience histological transformation. Transformation remains difficult to predict, and is often a harbinger of poor prognosis, especially in patients who are refractory to, or progress soon after prior treatment. Two types of tumor evolution patterns (standard linear and clonal branching/divergent) have been described as modes of transformation (Haebe et al., Blood, 2021). However, the molecular mechanisms responsible for these distinct pathways are poorly understood and traditional histopathologic testing methods have been suboptimal at identifying biologic models in real-world patients. Here, we describe the use of comprehensive molecular profiling, combining B-cell receptor (BCR) sequencing and clonal evolution assessment, to define different modes of histologic transformation in real-world cases.
Tissue biopsies and germline samples were collected from two reference cases before and after histologic transformation. Integrated genomic (whole-exome sequencing; WES) and whole transcriptomic (RNA sequencing; RNA-seq) analysis was performed using the BostonGene Tumor PortraitTM Test as previously described (Nuzdina et al., JCO, 2021). Computational assessment using BostonGene-developed algorithms was used to identify the molecular changes between the pre- and post-transformation samples.
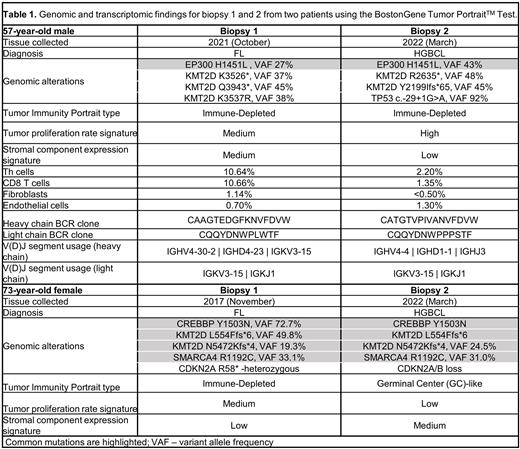
The first case involved a 57-year-old male diagnosed in 2021 with grade 1-2 FL. The patient received bendamustine and rituximab, but had progression with evidence of transformation six months later. A nodal biopsy showed a high-grade large B-cell lymphoma (HGBCL) with MYC and BCL-2 translocations and a proliferation rate greater than 99%. Comprehensive molecular profiling of lymph node biopsies from October 2021(FL) and March 2022 (HGBCL) showed that EP300 H1451L was the only common somatic mutation (Table 1). The mutational burden was higher in the HGBCL as compared to prior FL, and the genome presented triploidy and carried a TP53 splice site mutation. BCR analysis showed differences in the sequences of both heavy and light chains, as well as enrichment in specific clonotypes and V(D)J segments (Table 1). A common translocation in the IGH minor cluster region (MCR) was also found. Overall, these results support divergent clonal evolution of this tumor.
The second case involved a 73-year-old female with grade 1-2 FL on lymph node biopsy. Despite undergoing first-line immunochemotherapy with R-CHOP, she experienced disease recurrence and transformation to diffuse large B-cell lymphoma (DLBCL) three years later. The DLBCL sample showed clear features associated with aggressive disease, including expression of both MYC and BCL2. Comprehensive molecular profiling via integrated WES and RNA-seq analysis of lymph node biopsies from 2017 and 2022 revealed remarkable similarities between the two samples (Table 1). The only genomic difference was found in CDKN2A, which changed from a heterozygous mutation in the FL biopsy to a homozygous loss in the transformed sample. BCR sequencing showed enrichment in IgH and IgL clones with identical heavy chain variable region sequences and V(D)J segments used, suggesting that the 2017 and 2022 samples originated from the same tumor. The unchanged main clone in both samples suggests a standard linear transformation model, where the original FL clone and the transformed clone share common mutational and molecular profiles but show accumulation of additional deleterious genetic events in the latter.
In conclusion, the ability to perform clonal evolution analysis utilizing integrated genomic and transcriptomic methods can provide important information and differentiate emerging models of FL transformation, potentially leading to the identification of high-risk subsets and those who could benefit from alternative or targeted therapeutic approaches.
Disclosures
Almog:BostonGene: Current Employment, Current holder of stock options in a privately-held company, Divested equity in a private or publicly-traded company in the past 24 months; OncoDecipher, Inc: Consultancy. Novokreshchenova:BostonGene: Current Employment, Patents & Royalties. Smirnova:BostonGene: Current Employment. Brown:BostonGene: Current Employment, Current holder of stock options in a privately-held company. Kotlov:BostonGene: Current Employment, Current holder of stock options in a privately-held company, Patents & Royalties. Meerson:BostonGene: Current Employment. Anosov:BostonGene: Current Employment. Akpek:Pacific Central Coast Health Centers at Dignity Health: Current Employment. Ayrons:Mission Hope Medical Oncology: Current Employment, Current equity holder in private company. Fowler:Roche: Consultancy, Research Funding; Gilead: Consultancy, Research Funding; Celgene: Consultancy, Research Funding; BostonGene Corporation: Current Employment, Other: Leadership. Ferguson:Diagnostic Pathology Services of the Central Coast: Current Employment, Current equity holder in private company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal